Research
The Parks Lab is focused on addressing the question of how genetics and diet
interact together to contribute to common metabolic diseases, such as obesity,
diabetes, and cardiovascular disease. Through the use of large-scale systems
genetics studies we aim to identify and characterize biological pathways that
can be harnessed to treat disease. Our work is interdisciplinary in nature
and incorporates genetics, computational biology, molecular biology, and
biochemistry.
Our Current Projects
Molecular and physiological characterization of Agpat5

Agpat5 (1-acylglycerol-3-phosphate O-acyltransferase 5) was identified within a
genome-wide significant locus on chromosome 8 associated with plasma insulin
levels after 8 weeks of high-fat, high-sugar feeding. To test if Agpat5 is a
novel gene associated with plasma insulin levels and insulin resistance we
developed an antisense oligonucleotide (ASO) that silences Agpat5 expression
in vivo. In our published studies, we were able to demonstrate that silencing
Agpat5 with ASOs improves glucose tolerance in obese mice and rats2. We have
extended this observation and now have evidence that inhibiting Agpat5 in obese
mice improves the action of insulin in insulin resistant mice.
Agpat5 is not well studied and has never been implicated to play
a role in insulin resistance or diabetes. Biochemically, Agpat5 is a lipid
acyltransferase gene that can catalyze the conversion of lysophosphatidic acid
to phosphatidic acid. We are actively engaged in biochemical and molecular
studies of Agpat5 using both in vitro and in vivo model systems.
Network prioritization of human lipid GWAS loci
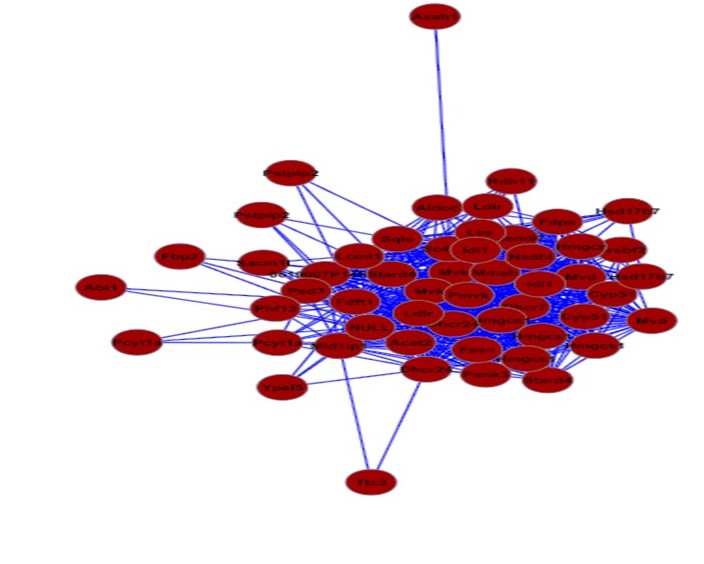
Large genome-wide association studies (GWAS) in humans have revolutionized our
ability to identify biological pathways and genes that influence complex traits
such as plasma cholesterol levels. However, identifing the causal gene within
genetic loci remains challenging. For many genome-wide significant loci, we
have yet to discover the underlying gene or mechanisms.
To aid in
prioritizing and identifying genes that contribute to variation in plasma
cholesterol, LDL, and HDL levels we are applying a suite of network-based
approaches to genome-wide gene expression and proteomic datasets that we and
others have generated. Through this approach we have already identified a highly
enriched sub-network of genes that are highly enriched for cholesterol
biosynthesis genes. We are actively engaged in building genome-wide networks and
screening candidate genes through in vitro and in vivo experiments.
Epistatic interactions in complex traits

Epistasis (non-additive gene-gene interactions) is ignored when performing
genome-wide association studies (GWAS). Epistasis can be detected using brute
force methods by performing pairwise association tests between a trait and
each pair of single nucleotide polymorphisms (SNPs). While computational
intensive, it is possible to test for epistasis across the genome. We are actively
optimizing methodology to exhaustively test for epistatic interactions in high
dimensional datasets. We have already identified statistically significant epistatic
interactions and plan to test if these interactions will replicate in independant
mouse populations
Defining the genetic nature of weightloss
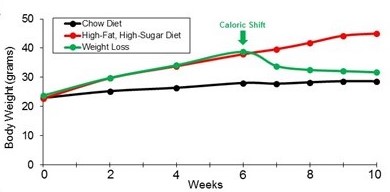
How does a dramatic shift in caloric intake contribute to weight balance? To address
if genetics regulates this process, we have initiated studies that assess how mice
respond to dramatic shifts in food caloric content by utilizing a mouse model for
this common form of dieting. Using this model, we have observed dramatic weight-loss
of mice after only 4 weeks of changing their diet to normal caloric diet. Current
work is engaged in determining how specific diets modulate this weight-loss.